library(tidyverse)
library(skimr)
library(gt)
library(ggthemes)
library(cowplot)
library(magick)30 Day Chart Challenge- Flora and Fauna
It is Day 3 of the #30DayChartChallenge. More info can be found at the challenge’s Github page. Today’s theme is flora and fauna. I found a trove of fascinating data at Global Assessment of Reptile Distributions. I chose the dataset on body size/ mass from the paper: “Different solutions lead to similar life history traits across the great divides of the amniote tree of life.” by Shai Meiri, Gopal Murali, Anna Zimin, Lior Shak, Yuval Itescu, Gabriel Caetano, and Uri Roll (Journal of Biological Research-Thessaloniki), 202128: 3.
Okay, let’s go. I’m going to keep the libraries to a minimum. That’s always my goal, but yet I ended up with 6!
fauna <- read_csv("animals.csv", show_col_types = FALSE)I’m going to change the class, clade, order, family and bionomial_2020 to factors. I’m keeping those columns and the mass and discarding the rest.
fauna_cleaned <- fauna %>%
select(Class:`body mass (g)`) %>%
select(-`binomial_(original files)`) %>%
rename(mass_g = `body mass (g)`, name = binomial_2020) %>%
mutate(Class = factor(Class),
Clade = factor(Clade),
order = factor(order),
family = factor(family),
name = factor(name))So, let’s see what kind of data we have.
table1 <- fauna_cleaned %>%
count(Class)
gt(table1)| Class | n |
|---|---|
| Aves | 9534 |
| Mammalia | 5840 |
| Reptilia | 11240 |
We have data on more than just reptiles, the dataset includes information about birds and mammals as well. But I’m only interested in reptiles.
reptiles <- fauna_cleaned %>%
filter(Class == "Reptilia")table2 <- reptiles %>%
count(Clade, order)
gt(table2)| Clade | order | n |
|---|---|---|
| Crocodylia | Crocodylia | 24 |
| Rhynchocephalia | Rhynchocephalia | 1 |
| Squamata | Squamata (Amphisbaenia) | 195 |
| Squamata | Squamata (Sauria) | 6868 |
| Squamata | Squamata (Serpentes) | 3837 |
| Testudines | Testudines | 315 |
Everyone knows that turtles are the best type of reptile, so let’s filter even further.
turtles <- reptiles %>%
filter(Clade == "Testudines")
table3 <- turtles %>%
count(order, family)
gt(table3)| order | family | n |
|---|---|---|
| Testudines | Carettochelyidae | 1 |
| Testudines | Chelidae | 53 |
| Testudines | Cheloniidae | 6 |
| Testudines | Chelydridae | 3 |
| Testudines | Dermatemydidae | 1 |
| Testudines | Dermochelyidae | 1 |
| Testudines | Emydidae | 47 |
| Testudines | Geoemydidae | 69 |
| Testudines | Kinosternidae | 24 |
| Testudines | Pelomedusidae | 18 |
| Testudines | Podocnemididae | 8 |
| Testudines | Testudinidae | 54 |
| Testudines | Trionychidae | 30 |
Let’s take a look at how big (or mighty, as some might say) the different families of turtles are. There is a very large range of masses so I’m using a log scale.
ggplot(turtles, aes(x = family, y = mass_g, color = family)) +
scale_y_log10() +
geom_boxplot() +
ggtitle("Mightiness of Different Families of Turtle and Tortoise") +
ylab("mass (g)") +
theme(legend.position = "none" ,
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1))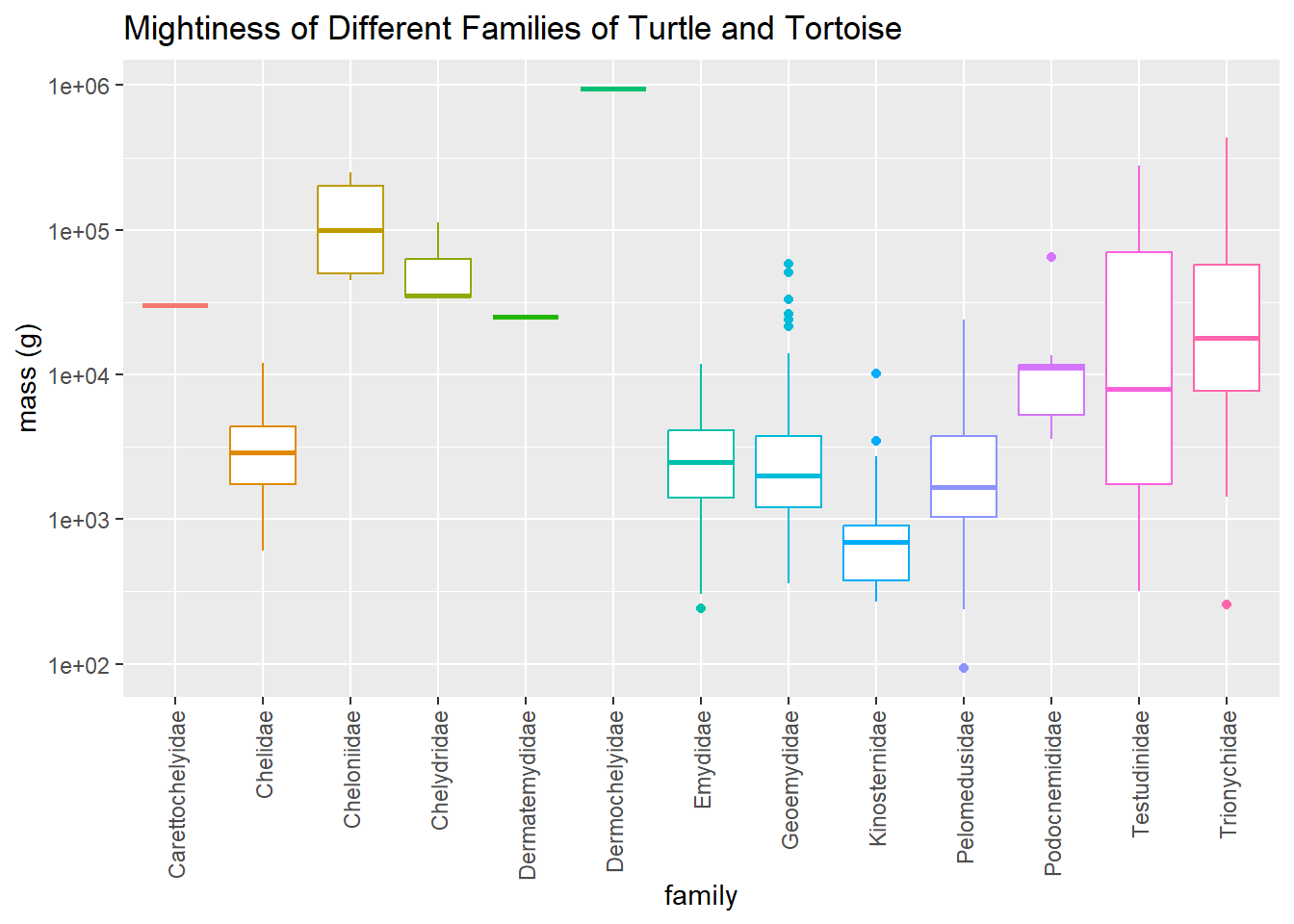
The heaviest turtles are the giantic leatherback turtles, a species of SEA TURTLE that weigh hundred of kg (around 1,000 lbs). The smallest turtle is the African dwarf mud turtle (Pelusios nanus), family Pelomedusidae, which full grown weighs under 100 grams.
Now let’s look at everyone’s favorite family of turtles, Emydidae. This family is often known as pond or marsh turtles. Filter them out, and add a category for Eastern Box Turtles. I called the variable box_turtle, but I’m only marking Eastern. There are other types of box turtles though not all of them are in Emydidae. Asian box turtle species were reassigned to the family Geoemydidae.
The common name box turtle, arises from the fact that these species have a hinge on the bottom shell, and can close up/ box up completely. Other turtles and tortoises can only pull their bits within their shell.

pond_turtles <- turtles %>%
filter(family == 'Emydidae') %>%
mutate(box_turtle = ifelse(name == "Terrapene carolina", TRUE, FALSE)) Okay, let’s look at mightiness of the turtles in this family.
turtle_plot <- pond_turtles %>%
ggplot(aes(x = fct_reorder(name, mass_g), y = mass_g, fill = box_turtle)) +
scale_fill_manual(values=c("#999999", "#E69F00")) +
geom_col(width = 0.7, position = position_dodge(10)) +
coord_flip() +
ylab("mass (g)") +
xlab("") +
ggtitle("Mightiness of Different Turtles in family Emydidae") +
labs(caption = "Data from https://doi.org/10.1186/s40709-021-00134-9") +
theme_classic() +
theme(axis.text = element_text(size = 6)) +
theme(legend.position = "none")
#found how to add an image to my graph on stack overflow
#https://stackoverflow.com/questions/63442933/how-can-i-add-a-logo-to-a-ggplot-visualisation
img <- image_read("pqtk5r.jpg")
# Set the canvas where you are going to draw the plot and the image
ggdraw() +
# Draw the plot in the canvas setting the x and y positions, which go from 0,0
# (lower left corner) to 1,1 (upper right corner) and set the width and height of
# the plot. It's advisable that x + width = 1 and y + height = 1, to avoid clipping # the plot
draw_plot(turtle_plot,x = 0, y = 0.15, width = 1, height = 0.85) +
# Draw image in the canvas using the same concept as for the plot. Might need to
# play with the x, y, width and height values to obtain the desired result
draw_image(img, x = 0.6, y = 0.35, width = 0.45, height = 0.45) 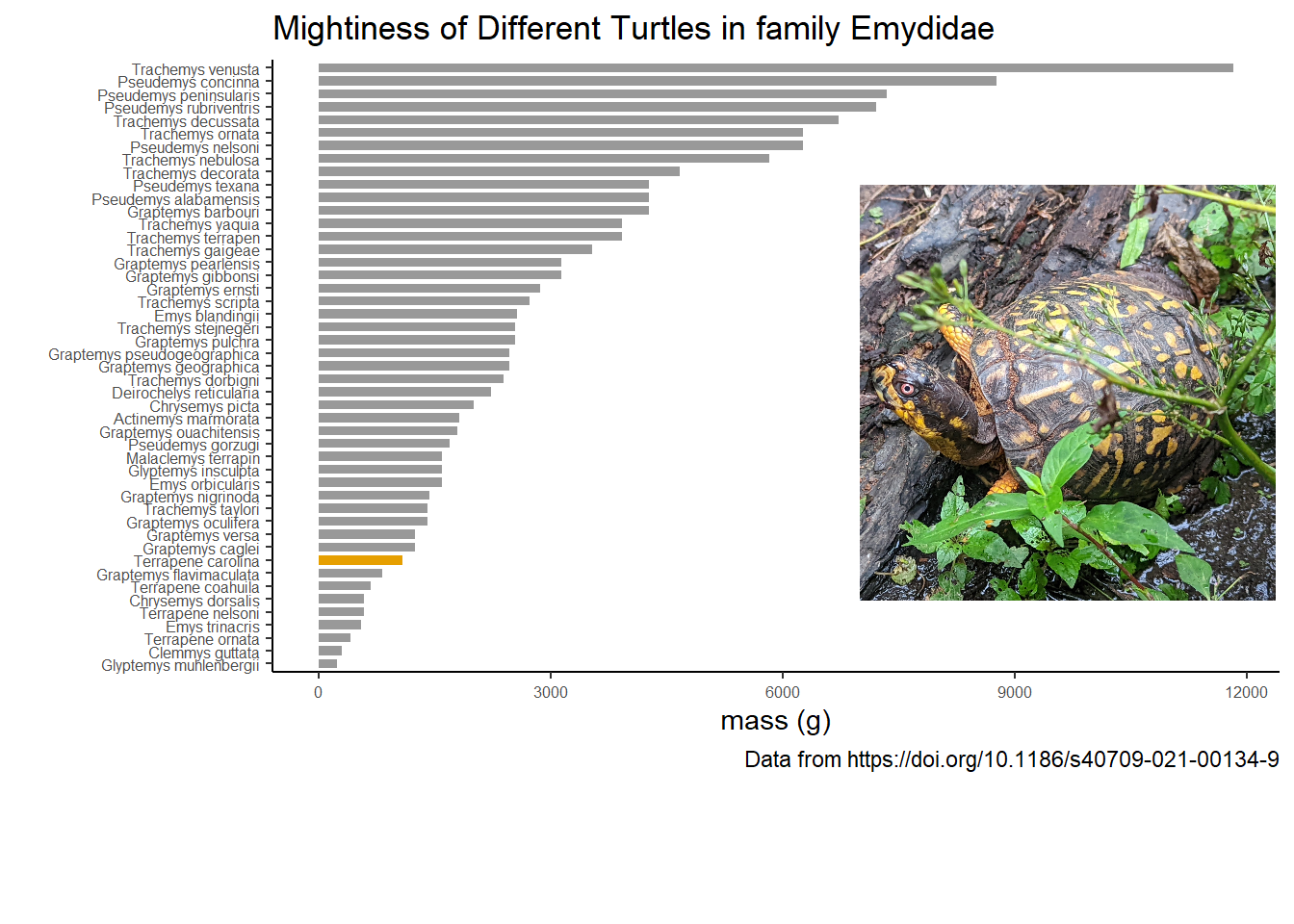
Citation
@online{sinks2023,
author = {Sinks, Louise E.},
title = {30 {Day} {Chart} {Challenge-} {Flora} and {Fauna}},
date = {2023-04-03},
url = {https://lsinks.github.io/posts/2023-04-03-chart-challenge-3/day3.html},
langid = {en}
}